Technology
Next Generation Sequencing big data processing.
To accelerate NGS big data processing Genalice has developed patented technology and implemented novel algorithms that produce highly accurate results.
Genalice technology is innovative and smart, giving us a unique position in the NGS arena. It is embodied in software that runs on commodity hardware.
We apply the principles of data reduction and data flow processing, without sacrificing quality to unlock the value that is hidden in the vast volumes of NGS data and deliver timely and accurate patient service.
The CPU is not the problem
Genalice technology exploits the fact that today’s processors have sufficient processing power to process large volumes of NGS data, provided it can readily access it. We ensure that data is transferred to the multi-core center of the processor and stays there until the parallel pipeline is done – using the full potential of the hardware. Current and next generation of commodity processors and memory systems have the right balance of compute and bandwidth to assure efficient processing of large volumes of NGS data using Genalice technology.
Secondly Genalice applies smart new algorithms that utilise the latest commodity hardware functionalities. This reduces the required processor cycles for NGS data processing dramatically. The combination of Data Flow Processing and Instruction Reduction boosts the performance beyond hardware acceleration of computational genomics solutions.
Thirdly we implement significant Data Stream Reduction, which applies to both memory and disk structures. This saves on disk space, being an important cost component, and reduces the required network and memory bandwidth. The approach assures optimal data flow within a node and between nodes for a HPC or Cloud environment.
Given that NGS data production expansion will far exceed the expected technology growth in CPU, network, and storage capacity, Genalice makes it possible to take NGS data usage to the next level. Now and in the future.
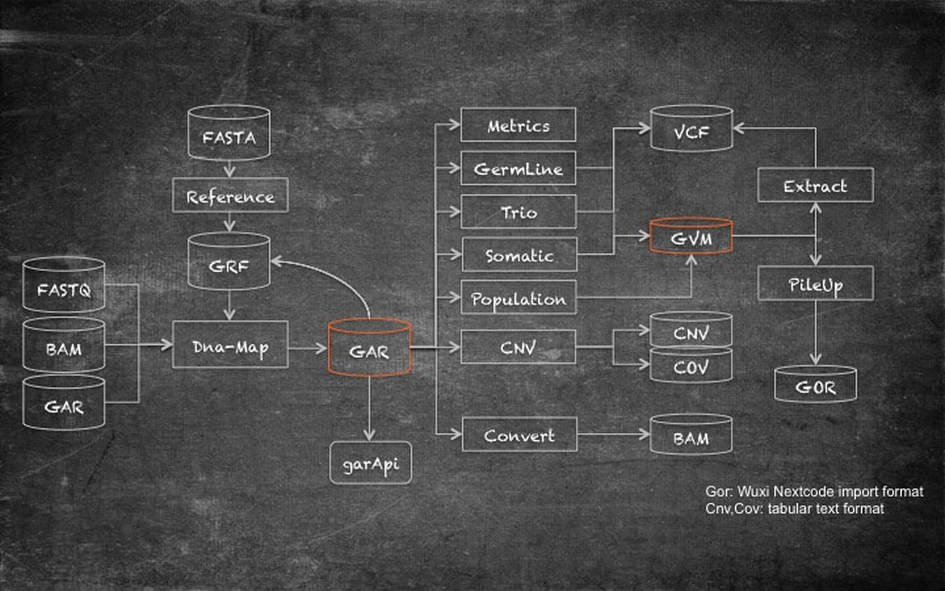
Secondary Analyis Workflow
Genalice solutions' monolithic functions all work together in an easy-to-use, comprehensive, expandable workflow. Read mapping can be done from different input formats (FASTQ, BAM, GAR). This makes it easy to align both newly sequenced and existing data e.g. for a migration of sample sets from GRCh37 to GRCh38. Genalice Map uses a reference (GRF) file that is compiled from a standard FASTA file. The FASTA file is translated to binary format for efficiency and unknown reference stretches are compressed. When the FASTA file has ALT sections, these can be tagged in the reference file to allow the aligner to adapt the scoring model when a choice must be made between reference and alternative section mapping. Read mapping with Genalice Map is a single step operation. All aligned reads are kept in memory and are written out in small footprint encoded GAR (Genalice Aligned Read) format at the end of the alignment run. Mate Pulling for Pair End data, coverage data collection and duplicate read registration are all part of the mapping process.
The GAR file is the common file from where all further analysis is done. The GAR file can be converted to BAM format, and the GAR file can be accessed directly by tools via the garAPI. The garAPI reports aligned reads in bamRecord format to the caller. This ensures seamless compatibility.
Patented variant calling technology, is at the heart of germline, somatic, trio and population calling. Next to producing a standard VCF file, we can store variants from multiple samples in a GVM. The GVM is a highly efficient variant repository that can store thousands of samples and hundreds of millions of variants. Content of the GVM can be extracted into a VCF file. This can be a single sample (g)VCF or a multi sample VCF. These files can become very large and can automatically be zipped.
Genalice CNV allows for comprehensive coverage and Copy Number Variation analysis. The results are stored in tabular text files with .COV and .CNV extension.
PileUp selects samples from GVM and creates a detailed output of coverage and un-optimised variants. These are stored in the Wuxi-NextCode proprietary GOR format which is used to import genetic data into the NextCode analysis platform.
Genalice ensures that having high performance tools are not overshadowed by reduced quality or increased complexity to operate. The solution is designed to improve data processing at the resource and at the usability level as it is clear that scaling up volumes in the NGS domain can only be done through performance increase and de-skilling the data processing steps.
Small Footprint aligned reads (GAR)
The GAR file (Genalice Aligned Reads) is equivalent to the BAM file, yet has a footprint of ±5GB for a full genome (30x) instead of the traditional 100GB. The GAR file holds meta-data, run-time statistics, quality metrics and pre-compiled coverage data. Storage footprint reduction is achieved using reference backed encoding and quality binning. Reducing the footprint saves on storage, writing time of the file, and transfer via network is much easier than with bigger files.

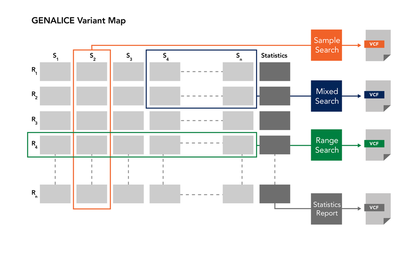
Scalable sample and variant repository (GVM)
Genalice Population is a unique tool implementing the well known concept of using intrinsic population observations to improve variant calling. This in context calling, or CBCE improves both sensitivity and precision. The implementation scales linear from a few to many samples and from one to many nodes.
The variants for a population or cohort are stored in a GVM (Genalice Variant Map). This is a repository that can be used to manage the samples and to search for patterns in the genetic profile. Using the GVM allows you to play with the data. You can take different groups of samples out of a large GVM into a separate GVM to further enhance the quality of the calls in e.g. a phenotype related cohort.
The speed and ease of use of Genalice Population frees up time to really focus on the meaning of the data instead of dealing with the data. The storage system is designed and tested to run with consistent file systems such as S3 from Amazon.
The signature of a GVM can be captured in a profile. This profile contains condensed information about the calls and their occurrences in a GVM. Such profile can be used to apply Consensus Based Call Enhancement during germline calling or somatic calling as if the samples were called as part of the GVM cohort the profile was derived from.
Unlike other solutions, Genalice Population is incremental. This means that adding 100 samples to a population of 1,000 takes the processing cost of only 100. There is no need to revisit the 1,000 samples that are already part of the GVM. This method allows dealing with thousands of samples in a linear way following a natural production flow.
There are no products listed under this category.
